1 Introduction
1.1 Background
A cooperative agreement [3] was signed between the Regenstrief Institute (RII) and the SNOMED International (IHTSDO) in July 2013. The agreement details how SNOMED CT and LOINC will be linked through:
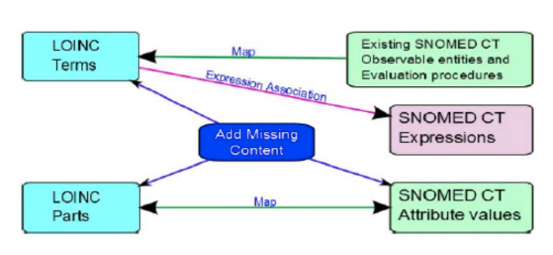
- a map of LOINC Terms to post-coordinated SNOMED CT expressions
- a map of LOINC Parts to SNOMED CT concepts
- a map between LOINC Terms and existing pre-coordinated SNOMED CT concepts
- LOINC Answer sets mapped to SNOMED CT codes
- addition of new content in either terminology
See Figure 1 for an illustration of these connections. This Production Release includes artifacts related to items 1, 2, and 5 above.
Figure 1. Diagram showing an overview of maps, Expression Associations and content additions that form part of the Cooperative Works of the Agreement [3].
1.2 Purpose
This document is intended to give a brief description, background context and explanatory notes on SNOMED International’s official RF2 release format of the LOINC - SNOMED CT Cooperative Project Production Release [1]. This is the format intended to be continually distributed for implementation.
Note that the information contained in the detailed Mapping and Modeling Guidelines section will be added to the SNOMED CT Editorial Guidance document.
This is not a detailed technical document about the release file formats, use cases, or on implementing LOINC and SNOMED CT together. See “Using LOINC with SNOMED CT” [2] for information about implementation of LOINC and SNOMED CT together.
All content will be released by both SNOMED International and the Regenstrief Institute through their distribution channels, e.g. SNOMED International Confluence site, LOINC website.
- Previous releases during the Alpha testing phase included two additional file formats (OWL and Excel) to assist in reviewing the content. Now that we are releasing the Production version, we will only publish the content in the official RF2 release format.
1.3 Scope
This document is written for the purpose described above and is not intended to provide details of the technical specifications for SNOMED CT or encompass every change made during the release.
1.4 Audience
The audience includes National Release Centers, vendors of electronic health records, terminology developers and managers who wish to partake in the review of the LOINC - SNOMED CT Cooperative project edition Production release.
2 Content Development Activity
2.1 Overview
This Production release contains two content files: a LOINC Term to SNOMED CT Expression Reference Set and a LOINC Part to SNOMED CT Reference Set. This Production release is RF2 compliant and it contains a full release file, a snapshot file and a delta file. The Production content is released under the 715515008 | LOINC - SNOMED CT Cooperation Project module (core metadata concept) with concept identifiers from the 20170731 version of the International release of SNOMED CT and Version 2.58 of LOINC released December 2016.
This release is based on an updated list of LOINC Parts received since the last Technology Preview (Alpha) Release. 3M assisted us in mapping additional LOINC Parts to SNOMED CT concepts since the last Alpha release. This release includes a majority of the Top 2000 LOINC Lab Observations and LOINC Parts needed to represent these LOINC Terms. In addition, many LOINC Parts not used in the Top 2000 LOINC Lab Terms are included in this release. Some content has been excluded from the Production Release:
We have not yet mapped all the LOINC Parts associated with the Top 2000 LOINC Terms to SNOMED CT concepts (or have not created the SNOMED CT concepts). The main reason for this exclusion is the requirement for further clarification with the Substance Project, RII, and Observable Model Redesign Project.
There are Terms being discussed internally or with Regenstrief for modelling reconsiderations and possible modification:
Susceptibility testing
Genetic testing
Terms defined by LP Components containing “tested for”
Terms containing disjunction
Physiological measurements and Vital signs Terms are not included because their scope is still being discussed with RII. Furthermore SNOMED International has not received the list of LOINC Parts from RII used to define these Terms.
Veterinary specific terms are not considered in the scope of SNOMED CT and this project
Although panel names are in the scope of the Cooperative Agreement, modeling of panel Terms is not. However, SNOMED International decided not to include the LOINC Term with panel names.
2.2 Content usage
The main purposes for the mapping are to (a) link the laboratory coverage of LOINC with the SNOMED CT concept model to provide a consistent model rather than having divergent representation of similar concepts, (b) limit duplication of effort related to overlapping areas, and (c) focus resources on common and collaborative effort rather than the competing activities of the past. In practical terms there are a variety of ways to deploy and benefit from the resulting products. These are explored in the current draft of the document “Using LOINC with SNOMED CT.” It is available on Confluence at http://snomed.org/snomedloinc.
2.3 Content detail
2.3.1 LOINC Term to SNOMED CT Expression Reference Set
The LOINC Term to SNOMED CT Expression Reference Set contains 21971 active LOINC Terms associated with post-coordinated SNOMED CT expressions. This refset includes the majority of the LOINC Terms from the Top 2000 Laboratory LOINC list.
All of the LOINC Terms are subclasses of 363787002 | Observable entity (observable entity) in SNOMED CT and the expressions follow the latest version of the SNOMED CT Observables model. [4,5,6].
See Table 1 for an example from the refset and see Figure 2 for a diagram of the Observables model used in this work.
Section “Mapping and Modeling Guidelines” contains general rules on the mapping and exceptions.
The majority of the expressions included are fully defined and are an exact match with a LOINC Term, with some exceptions.
The expressions are represented in the stated form with no hierarchy included
There are some known content issues in this reference set which are under discussion with the Observables and Investigation Modeling Project Group and Regenstrief Institute.
- See the SNOMED CT Technical Implementation Guide for more information about Expressions [7]
A.
id | mapTarget | expression | correlationId |
731850f9-3dbd-4f30-81bf-ee5ab5223154 | 27182-5 | Calcium:PrThr:Pt:Bld:Ord | 363787002|Observable entity|:704319004|Inheres in|=87612001|Blood|,704327008|Direct site|=119297000|Blood specimen|,370134009|Time aspect|=123029007|Single point in time|,246093002|Component|=259295006|Calcium electrolyte|,370132008|Scale|=117363000|Ordinal value|,704318007|Property type|=705057003|Presence| | 447557004 | Exact match map from SNOMED CT source code to target code (foundation metadata concept) |
B.
id | mapTarget | expression | correlationId |
731850f9-3dbd-4f30-81bf-ee5ab5223154 | 27182-5 | 363787002:704319004=87612001,704327008=119297000,370134009=123029007,246093002=259295006,370132008=117363000,704318007=705057003 | 447557004 |
Table 1. Example from the LOINC Term to SNOMED CT Expression Reference Set. This shows a LOINC term “Calcium:PrThr:Pt:Bld:Ord” (LOINC code 27182-5) with an equivalent post-coordinated expression. Note that “A” includes text labels for demonstration purposes; the actual refset will not contain the text descriptions (shown in “B”). For brevity, not all fields in the LOINC Term to SNOMED CT Expression Reference Set are included in this table.
A.
B.
Figure 2. Observables model showing the relationship of SNOMED CT attributes (in yellow) and values (in blue) used in creating the post-coordinated expressions in the LOINC Term to SNOMED CT Expression Refset. “A” shows the model for quality observables and “B” shows the model for process observables. Note that all the permissible value hierarchies are not shown in this diagram for brevity. See 6.3.3 Attributes for observable entities and evaluation procedures [6] in the SNOMED CT Editorial Guide for a complete list of approved hierarchies for values of each attribute.
2.3.2 LOINC Part to SNOMED CT Map Reference Set
The LOINC Part to SNOMED CT Reference Set consists of 6629 LOINC Parts (LP) mapped to one or more SNOMED CT concepts.
While the majority of the maps are one to one, occasionally one LP maps to more than one SNOMED CT concept or a combination of LPs map to one SNOMED CT concept or combination of SNOMED CT concepts. Additionally, a concept may be used as a value in more than one attribute.
See section “Mapping and Modeling Guidelines” for general rules on the mapping and exceptions.
- See Table 2 for examples of Part maps.
A.
id | mapTarget | referencedComponentId | attributeId | correlationId |
2a1d16c5-f45d-4758-8828-dc8021f86711 | LP15257-6 | Calcium | 259295006 | Calcium electrolyte (substance) | 246093002 | Component (attribute) | 447559001 | Broad to narrow map from SNOMED CT source code to target code (foundation metadata concept) |
76c69030-5b1d-4013-8e8d-26288ad5f9ff | LP15257-6 | Calcium | 259295006 | Calcium electrolyte (substance) | 704324001 | Process output (attribute) | 447559001 | Broad to narrow map from SNOMED CT source code to target code (foundation metadata concept) |
B.
id | mapTarget | referencedComponentId | attributeId | correlationId |
2a1d16c5-f45d-4758-8828-dc8021f86711 | LP15257-6 | 259295006 | 246093002 | 447559001 |
76c69030-5b1d-4013-8e8d-26288ad5f9ff | LP15257-6 | 259295006 | 704324001 | 447559001 |
Table 2. Example of one LOINC Part mapped to a SNOMED CT concept to be used with two different attributes. LP15257-6 is a component part type in LOINC. The attributeId column indicates for which attribute in SNOMED CT this concept would be a value; in this case the concept is used as value for two different SNOMED CT attributes. Note that A includes text labels for demonstration purposes; the actual refset will not contain the text descriptions (shown in B). For brevity, not all fields in the LOINC Part to SNOMED CT Complex Map Reference Set are included in this table.
3 Mapping and Modeling Guidelines
3.1 General Guidelines
Mapping is performed from LOINC to SNOMED CT
As part of the LOINC - SNOMED CT Cooperation Project agreement, LOINC Terms are being defined with post-coordinated expressions using the Observable entity model
The list of attributes and values used in the SNOMED CT observable entity model is available in section 6.3.3 Attributes for observable entities and evaluation procedures [6] in the SNOMED CT Editorial Guide.
In this Production Release we have modeled LOINC Terms as either a quality observable or a process observable. Definitions of these, provided in the Observables and Investigation procedures redesign Inception/Elaboration document [5], are below. See Figure 2 for a list of the attributes used to model quality and process observables.
Quality observable
An observable about a characteristic or feature, a quality, that is inherent in a person or a thing. Examples include the mass of a human being and the concentration of sodium in plasma.
Rate observables about the amount of output of a process (e.g., the volume of urine voided during a time interval, the volume of blood plasma that is cleared of creatinine per unit of time) are modeled as quality observables.
Process observable
- An observable about a feature of a process or specific outcome of a process. Examples include durations and various kinds of rates such as secretion rate.
3.2 LOINC Part to SNOMED CT Concept and Attribute Alignment
In general, we mapped from LOINC axis to SNOMED CT attribute as such:
LOINC Component to SNOMED CT attributes
LOINC Component → 246093002 | Component (attribute) for quality observables. See Figure 2A for a list of the attributes used to model quality observables.
id | mapTarget | referencedComponentId | attributeId | correlationId |
3ffe4c79-7498-4c46-86df-e61cea435e16 | LP16819-2 | Formaldehyde | 111095003 | Formaldehyde (substance) | 246093002 | Component (attribute) | 447557004 | Exact match map from SNOMED CT source code to target code (foundation metadata concept) |
LOINC Component → 704324001 | Process output (attribute) and 704321009 | Characterizes (attribute) (and optionally 704322002 | Process agent (attribute)) in process observables such as excretions. See Figure 2B for a list of the attributes used to model quality observables.
id | mapTarget | referencedComponentId | attributeId | correlationId |
9ffba7b6-dd03-4d09-9f2d-d8d8c5390d4a | LP15141-2 | Silver | 41967008 | Silver (substance) | 704324001 | Process output (attribute) | 447558009 | Narrow to broad map from SNOMED CT source code to target code (foundation metadata concept) |
3ec0cccf-1f72-4844-8778-88536ee36ef6 | LP15141-2 | Silver | 718500008 | Excretory process (qualifier value) | 704321009 | Characterizes (attribute) | 447558009 | Narrow to broad map from SNOMED CT source code to target code (foundation metadata concept) |
0503a564-f8c1-40ce-a174-92f4282a2ecc | LP15141-2 | Silver | 64033007 | Kidney structure (body structure) | 704322002 | Process agent (attribute) | 447558009 | Narrow to broad map from SNOMED CT source code to target code (foundation metadata concept) |
When the LOINC Component contains additional information/qualifiers, we mapped as such:
LOINC Components containing divisors → 246093002 | Component (attribute) + 704325000 | Relative to (attribute)
LOINC Components containing challenges → 246093002 | Component (attribute) + 704326004 | Precondition (attribute)
LOINC Components containing adjustments → 246093002 | Component (attribute) + 246501002 | Technique (attribute)
Sometimes a new SNOMED CT concept containing a combination of techniques was needed
Exceptions, special cases and further information
We do not map the LOINC Components Appearance, Color, and Number to SNOMED CT concept. We instead use an appropriate SNOMED CT value from the property subhierarchy in the 704318007 | Property type (attribute).
Observables entities are not allowed as values for the attributes in the observable entity model (and therefore LOINC post coordinated expression). When a LOINC Component is an observable entity, it should be mapped to the actual analyte being measured. Mapping to extra attributes (e.g. technique) might become required. Some examples
Estimated Glomerular filtration rate, MDRD formula (eGFR)
RBC sedimentation rate
Anion gap, Anion gap 3, and Anion gap 4
Base excess or Base deficit
For LOINC Parts that are anions and cations, we map to the electrolyte concept in SNOMED CT. The generic substance concept is considered to be a grouper for the electrolyte concept. See Table 2 for an example of a mapping.
Substance LOINC Components are in general aligned with concepts in the substance hierarchy rather than the product hierarchy in SNOMED CT. Concepts in the product hierarchy are defined by substance concepts via the “Has active ingredient (attribute)." In the measurement of drugs (or any product), what is actually being measured is the active ingredient. So while the range of “Component (attribute)” includes both products and substances, we align with substance hierarchy concepts in the vast majority of cases.
Some LOINC Components include “X organism species” such as “Salmonella species…” The use of terms of the form "X species" is routine in laboratory reporting, but in the context of the Linnaean organism hierarchy there is no difference between "Salmonella species" and simply "Salmonella" the genus. In the context of a laboratory report, the term "Salmonella species" sometimes is intended to convey additional information beyond the place of the identified organism in the Linnaean hierarchy, but the intended connotation may vary from lab to lab and from organism to organism. Since the organism code represents a class of organisms, it cannot also represent what was or was not done, or what will be done, to identify the organism. It also cannot properly represent other information about the result. If there is additional information that needs to be communicated, it should be in a separate statement or comment (e.g. "further species identification pending" or "sent to reference laboratory for further identification," or "further identification to be done if clinically indicated.")
From IUPAC (International Union Of Pure And Applied Chemistry) Gold Book: "Nor" used to denote the elimination of one methylene group from a side chain of a parent structure (including a methyl group). "Desmethyl" means the removal or absence of a methyl group from the parent compound. The LOINC User's Guide section 2.1.2.20: For drug metabolites we will use the "Nor" rather than the "Desmethyl", e.g., for instance Nordoxepin not Desmethyldoxepin. There are no desmethyl% LOINC parts due to this approach.
Sometimes the LOINC Component also maps to 246501002 | Technique (attribute) in SNOMED CT. Example: LOINC Component Part Crystals.unidentified maps to 246093002 | Component (attribute) of "Crystal" and 246501002 | Technique (attribute) = "Detecting by light microscopy without classifying"
LOINC Components containing an organism name and a form of typing (e.g., serotype, phagetype, biotype) will be mapped as follows:
Examples: X organism serotype, X organism phagetype, X organism biotype
246093002 | Component (attribute) = X organism
246501002 | Technique (attribute) = serotyping, bacteriophage typing, biotyping, etc
Mapping for HLA antigens, alleles and cells.
LOINC Components containing “CDX” should be aligned with cell concepts, e.g., LP17730-0 "Cells.CD33" aligns with 116825007 | Cell positive for CD33 antigen (cell).
LOINC Components containing “CDX Ag” should be aligned with antigen substance concepts, e.g. LP LP37563-1 "CD3" maps to 44706009 | Lymphocyte antigen CD3 (substance).
LOINC Components that begin with HLA and include an asterisk are mapped to allele concepts.
As defined in the LOINC Users' Guide, "Peak" in LOINC refers to 'The time post drug dose at which the highest drug level is reached (differs by drug)' and "Trough" in LOINC refers to 'The time post drug dose at which the lowest drug level is reached (varies with drug).' Therefore these LPs are mapped to 703764006 | At time of peak level (qualifier value) and 703765007 | At time of trough level (qualifier value), respectively, as they are only used in combination with substances. For example, the LP Component "Gentamicin^trough" includes the Component "Gentamicin" and a Challenge of "Trough." In SNOMED CT this would be modeled as 246093002 | Component (attribute) = Gentamicin (substance) and 704326004 | Precondition (attribute) = At time of trough level (qualifier value).
LOINC Property to SNOMED CT attribute
LOINC Property → 704318007 | Property type (attribute)
LOINC Scale to SNOMED CT attribute
LOINC Scale → 370132008 | Scale type (attribute)
LOINC System to SNOMED CT attributes
LOINC System → 704327008 | Direct site (attribute) + 704319004 | Inheres in (attribute) (except when the LOINC Term has a property type of PRID)
Direct site is the equivalent of system in LOINC. Used for all specimen types except XXX.
Inheres in applies to all specimen types except “specimen”, XXX, Body fluid, tissue and aspiration specimen
Direct site and Inheres in definitions
The Direct site is the direct object of the observation action; the site or specimen where the observation takes place. This may be different from the value of Inheres in, since the property being observed may be inferred from a specimen or from a remote site. The intended object of the observation is the entity in which the observed property inheres; or the independent continuant which is ideally intended to be observed.
Example:
Serum specimen being used to determine body plasma sodium concentration. The concentration inheres in the plasma, but is being measured in a serum specimen.
Example mapping:
id | mapTarget | referencedComponentId | attributeId | correlationId |
73c6bd67-5a68-48dd-b48a-55a382d4e731 | LP 7494-0 | PPP | 119362004 | Platelet poor plasma specimen (specimen) | 704327008 | Direct site (attribute) | 447558009 | Narrow to broad map from SNOMED CT source code to target code (foundation metadata concept) |
6aa7669a-f95c-4526-bdb5-f20d1d654a52 | LP 7494-0 | PPP | 50863008 | Plasma (substance) | 704319004 | Inheres in (attribute) | 447558009 | Narrow to broad map from SNOMED CT source code to target code (foundation metadata concept) |
id | mapTarget | referencedComponentId | attributeId | correlationId |
9dc5c0b8-2d57-48fc-b487-1468721ccca9 | LP7370-2 | Isolate | 386126001 | Microbial isolate (substance) | 718497002 | Inherent location (attribute) | 447558009 | Narrow to broad map from SNOMED CT source code to target code (foundation metadata concept) |
309ab906-6a1d-4f4c-93ed-2218ce11b52d | LP7370-2 | Isolate | 119303007 | Microbial isolate specimen (specimen) | 704327008 | Direct site (attribute) | 447558009 | Narrow to broad map from SNOMED CT source code to target code (foundation metadata concept) |
Note:
When the LOINC Term has a property of PRID and scale of NOM --> property inheres in organism (named in LOINC component)
LOINC Time to SNOMED CT attributes
For quality observables:
LOINC Time Part Pt → 370134009 | Time aspect (attribute) = Single point in time in SNOMED CT
LOINC Time Part of X hours → 370134009 | Time aspect (attribute) = X hours in SNOMED CT
For process observables:
LOINC Time Part of X hours → 704323007 | Process duration (attribute) = X hours in SNOMED CT
LOINC Method to SNOMED CT attribute
LOINC Method → 246501002 | Technique (attribute)
Some LOINC Terms contain references to techniques in both the LP Component name and the LP Method name. In some cases we have used a SNOMED CT technique concept which represents the LP Method and technique from the LP Component name or adjustment name.
LOINC Units
LOINC Units → 246514001 | Units (attribute) in few cases
LOINC Units are example units or submitted units, and are not part of the 5-6 essential LOINC Parts that create LOINC Terms. Units of measure may change based on the setting and equipment, and can be standard or American units. We don't plan to map LOINC units unless they are absolutely required (for example, percentage was required to represent 100 cells). Percentage is understood and used the same everywhere.
3.3 Correlation between LOINC Part and SNOMED CT Concept and Attributes
Our goal is to create maps that are exact matches between LOINC Parts and SNOMED CT concepts. Where this is not possible, the map is given a designation other than “Exact match.”
Example: the LOINC Part “Leukocyte other” is mapped to “Leukocyte (cell)” and “Population of leukocytes in portion of fluid” with a correlation of “SNOMED CT Broader.”
Example: the LOINC Part “Beta lactamase.usual” is mapped to beta-Lactamase (substance) but this is not an exact match. However, since Beta lactamase.usual does not meet the inclusion criteria for SNOMED CT, we keep the map to the more general concept and label it as "LOINC Narrower/SNOMED CT Broader." LOINC Terms defined by this concept will be marked as primitive.
Example: We do not consider LOINC Parts with “NOS” equal to SNOMED CT concepts without “NOS.” “NOS” is specifically out of scope of SNOMED CT and would not be added. Therefore, the map from LOINC Parts with NOS to SNOMED CT concepts will be “LOINC narrower.”
Example: LOINC Parts with “tissue” or “body fluid” will be “LOINC narrower.” Body fluid System really means fluids that are not otherwise defined in LOINC, and not a higher-level concept that encompasses all body fluids. Tissue LOINC Parts are defined similarly in LOINC.
We mapped from one LOINC Part to one SNOMED CT concept where possible. In some cases a one to one map was not possible:
LOINC Part mapped to more than one SNOMED CT concept
LOINC System Parts are mapped using two attribute-value pairs in SNOMED CT: 704327008 | Direct site (attribute) and 704319004 | Inheres in (attribute)
Example: LOINC Part “Ser/Plas” is mapped to SNOMED CT 704327008 | Direct site (attribute) = Acellular blood (serum or plasma) specimen and 704319004 | Inheres in (attribute) = Plasma
LOINC Components containing typing will be mapped to an appropriate value in the 246093002 | Component (attribute) as well as to a typing technique concept such as biotyping, serotyping, etc. in 246501002 | Technique (attribute). If the LOINC Term is also defined by a Method in LOINC, a combination concept for the technique will be created and utilized.
LOINC Parts for cells (e.g. Basophils) potentially align with two concepts in SNOMED CT: an individual X cell concept and a population of X cells in portion of fluid concept. The property type and scale of the associated LOINC Term will determine which map is appropriate for that LOINC Term. For example, the LP "Basophils" could align with 246093002 | Component (attribute) = 702963004 | Population of all basophils in portion of fluid (body structure) and/or 246093002 | Component (attribute) = 30061004 | Basophil, segmented (cell).
id | mapTarget | referencedComponentId | attributeId | correlationId |
2078251f-30c3-4098-8044-a175d19a66ef | LP14328-6 | Basophils | 702963004 | Population of all basophils in portion of fluid (body structure) | 246093002 | Component (attribute) | 447558009 | Narrow to broad map from SNOMED CT source code to target code (foundation metadata concept) |
9acce8eb-7cc8-47ad-8ef2-7d01c7f4eb0b | LP14328-6 | Basophils | 30061004 | Basophil, segmented (cell) | 246093002 | Component (attribute) | 447558009 | Narrow to broad map from SNOMED CT source code to target code (foundation metadata concept) |
Some LOINC Component Parts (generally substances) are mapped using the 246093002 | Component (attribute) and/or 704324001 | Process output (attribute) depending on the time aspect and property of a LOINC Term. When the Property type is MRat, SRat, VRat, and NRat, and Time = X hrs, then the 704324001 | Process output (attribute) carries the SNOMED CT concept equivalent to the LOINC Component. Otherwise, the 246093002 | Component (attribute) carries the SNOMED CT concept equivalent to the LOINC Component. A LOINC Term will generally only include a value for 246093002 | Component (attribute) OR for 704324001 | Process output (attribute) but not both. The Part map may include a value for 246093002 | Component (attribute) and 704324001 | Process output (attribute).
Example: Creatinine is used as a value of the 246093002 | Component (attribute) in LOINC Term Creatinine/Calcium [Mass Ratio] in Urine. Creatinine is used as a value of the 704324001 | Process output (attribute) in LOINC Term Creatinine [Mass/volume] in 24 hour Urine.
LOINC Divisor Parts are mapped to two attribute-value pairs in SNOMED CT: 704325000 | Relative to (attribute) and 246514001 | Units (attribute)
LOINC Part 100 Leukocytes is mapped to SNOMED CT 704325000 | Relative to (attribute) = Population of all leukocytes in portion of fluid and 246514001 | Units (attribute) = Percentage
LOINC Part is not mapped to an equivalent SNOMED CT concept
LOINC Part that is not universal and reproducible is mapped to a more general concept in SNOMED CT.
Example, the LOINC Part “Leukocyte other” is mapped to “Leukocyte (cell)” and “Population of leukocytes in portion of fluid” in SNOMED CT because the meaning of “Leukocyte other” cannot be specified.
LOINC Part mapped to a different attribute in SNOMED CT concept
Example: The LOINC Component Part “Color” is mapped to the 704318007 | Property type (attribute) with a value of “Color” in SNOMED CT
Two LOINC Parts are combined to map to two SNOMED CT concepts
LOINC Component Part Crystals.unidentified and LOINC Method Microscopy.light map to SNOMED CT 246093002 | Component (attribute) = Crystal and 246501002 | Technique (attribute) = Detecting by light microscopy without classifying
3.4 LOINC Term to SNOMED CT Post Coordinated Expression Alignment
Our goal is to create a SNOMED CT expression equivalent to each LOINC Term and mark it as fully defined with a designation of “Exact match.” Where this is not possible, the map is marked as primitive and/or a designation other than “Exact match” is utilized. In this Production Release, the only correlation beside “exact match” used is 447559001 | Broad to narrow map from SNOMED CT source code to target code (foundation metadata concept), i.e. the SNOMED CT expression is broader than the LOINC Term. To date, we have not created an expression that is narrower than the LOINC Term, and it is unlikely that we ever would.
LOINC Terms that utilize the following LOINC Parts will be marked as primitive because the LOINC Part will not have an exact match in SNOMED CT.
LP7641-6 Tiss
LP189537-6 XXX.tissue
LP61649-7 Tissue
LP7642-4 Tiss.FNA
LP29168-9 Soft tissue.FNA
LP7037-7 Asp
LP7593-9 Specimen
Example: the LOINC Term “Leukocytes other [#/volume] in Body fluid” is mapped to an expression that has 246093002 | Component (attribute) = Population of leukocytes in portion of fluid along with other attribute-value pairs. The map is given a correlation of “LOINC narrower” and a designation of “Primitive.”
Example: the LOINC Part “Beta lactamase.usual” is mapped to beta-Lactamase (substance) but this is not an exact match. However, since Beta lactamase.usual does not meet the inclusion criteria for SNOMED CT, we keep the map to the more general concept and label it as "LOINC Narrower/SNOMED CT Broader." LOINC Terms defined by this concept will be marked as primitive.
Example: We do not consider LOINC Parts with “NOS” equal to SNOMED CT concepts without “NOS.” “NOS” is specifically out of scope of SNOMED CT and would not be added. Therefore, when linking the related LOINC Terms to SNOMED CT expressions, we set the definition status for the expressions containing "NOS" to “primitive.”
An expression only needs to include the attributes necessary to specify the LOINC Term. It does not need to use every attribute in the Observables model.
The LOINC Terms in this Production Release are modeled as either quality observables or process observables. See an example of a quality observable expression in the first row in the table and a process observable in the second row of the table. See definitions of quality and process observables in the General Guidelines section.
id | mapTarget | expression | correlationId |
b14bd80e-d892-4902-a18e-3039223460f3 | 5574-9 | Aluminum:MCnc:Pt:Ser/Plas:Qn | 363787002|Observable entity|:704318007|Property Type|=118539007|Mass concentration|,370134009|Time aspect|=123029007|Single point in time|,704327008|Direct Site|=122592007|Acellular blood (serum or plasma) specimen|,246093002|Component|=12503006|Aluminum|,370132008|Scale|=30766002|Quantitative|,704319004|Inheres In|=50863008|Plasma| | 447557004 | Exact match map from SNOMED CT source code to target code (foundation metadata concept) |
69617976-aad4-4baa-bcfe-2455daa7b0f7 | 15047-4 | Creatine:SRat:24H:Urine:Qn | 363787002|Observable entity|:704323007|Process Duration|=123027009|24 hours|,704321009|Characterizes|=718500008|Excretory process|,704322002|Process Agent|=64033007|Kidney|,704324001|Process Output|=14804005|Creatine|,370132008|Scale|=30766002|Quantitative|,704318007|Property Type|=118562009|Substance rate|,704327008|Direct Site|=122575003|Urine specimen| | 447557004 | Exact match map from SNOMED CT source code to target code (foundation metadata concept |
LOINC Terms with Presence vs. Presence Identification: X in Y
PRID: e.g. 14475-8 | Bacteria identified in Cervix by Aerobe culture
Bacteria → 704319004 | Inheres in (attribute)
Cervix → 718497002 | Inherent location (attribute)
id | mapTarget | expression | correlationId |
ecc8d499-7385-48d0-b632-210017722914 | 14475-8 | Bacteria identified:Prid:Pt:Cvx:Nom:Aerobic culture | 363787002|Observable entity|:704319004|Inheres In|=409822003|Superkingdom Bacteria|,370134009|Time aspect|=123029007|Single point in time|,370132008|Scale|=117362005|Nominal value|,246501002|Technique|=703750006|Aerobic culture|,718497002|Inherent Location|=71252005|Cervix uteri structure|,704318007|Property Type|=118584009|Presence OR identity|,704327008|Direct Site|=119395005|Specimen from uterine cervix| | 447557004 | Exact match map from SNOMED CT source code to target code (foundation metadata concept) |
704319004 | Inheres in (attribute) and 718497002 | Inherent location (attribute) are required in pair (except when specimen is XXX, aspiration, specimen, tissue, soft tissue).
Presence: e.g. 21262-1 | Escherichia coli shiga-like [Presence] in Stool by Immunoassay
Escherichia coli shiga-like → SNOMED CT Component
Stool → SNOMED CT Inheres In
Note: Identity (or type or taxon) is a property of the bacteria while presence is a property of the system where the bacteria were found. For example, the type of bacteria might be Escherichia coli but it's a property of the abscess to contain or not contain bacteria.
Modeling of Time in LOINC Terms
LOINC Terms modeled as quality observables will utilize the Time aspect attribute to represent time with a value of Single point in time, 24 hours, 2 hours, etc.
LOINC Terms modeled as process observables will utilize the Process duration attribute to represent time with a value of X hours.
See the table below for examples of expressions modeled as process observable (first row) and quality observables (second and third rows).
id | mapTarget | expression | correlationId |
5f4ad73f-8b24-432d-8242-4af8739573ac | 6931-0 | Silver:MRat:24H:Urine:Qn | 363787002|Observable entity|:704323007|Process Duration|=123027009|24 hours|,704321009|Characterizes|=718500008|Excretory process|,704322002|Process Agent|=64033007|Kidney|,704318007|Property Type|=118544000|Mass rate|,370132008|Scale|=30766002|Quantitative|,704324001|Process Output|=41967008|Silver|,704327008|Direct Site|=122575003|Urine specimen | 447557004 | Exact match map from SNOMED CT source code to target code (foundation metadata concept) |
159cc296-1e9c-452a-9d7c-44b70227d976 | 29977-6 | Silver:MCnc:24H:Urine:Qn | 363787002|Observable entity|:704318007|Property Type|=118539007|Mass concentration|,370134009|Time aspect|=123027009|24 hours|,246093002|Component|=41967008|Silver|,704327008|Direct Site|=122575003|Urine specimen|,370132008|Scale|=30766002|Quantitative|,704319004|Inheres In|=78014005|Urine| | 447557004 | Exact match map from SNOMED CT source code to target code (foundation metadata concept) |
2b5eb8a6-3ef3-4795-9c05-e59e12115137 | 9437-5 | Methylhippurate:MRat:Pt:Urine:Qn | 363787002|Observable entity|:246093002|Component|=115388007|Methylhippurate|,370134009|Time aspect|=123029007|Single point in time|,704318007|Property Type|=118544000|Mass rate|,370132008|Scale|=30766002|Quantitative|,704319004|Inheres In|=78014005|Urine|,704327008|Direct Site|=122575003|Urine specimen | 447557004 | Exact match map from SNOMED CT source code to target code (foundation metadata concept) |
If the expressions are classified, some more specific LOINC Terms are subsumed by more general LOINC Terms. We have verified the validity of the following subsumptions with Regenstrief Institute and the Observables and Investigation Model Project Group:
Subsumption of expressions due to SNOMED CT substance hierarchy organization:
Expressions containing SNOMED CT “Amino-acids” concepts subsume individual amino acid
Expressions containing SNOMED CT “Antibody” concepts subsume individual antibodies
Expressions containing SNOMED CT substances which include free, soluble, or unsaturated substances are subsumed by the generic substance concept
Expressions containing SNOMED CT substances which include organ/body part (e.g. Alkaline phosphatase.renal) are subsumed by the more general substance concept
Expressions for LOINC Terms with a technique are subsumed by expressions with a more generic method or methodless expressions
Expressions for LOINC Terms containing adjustments are subsumed by generic or unadjusted expressions
Expressions for LOINC Terms containing challenges are subsumed by generic expressions
Expressions for LOINC Terms with a more specific specimen are subsumbed by more generic expressions, e.g. Arterial blood is necessarily a kind of Blood
Expressions for LOINC Terms with a more specific property are subsumed by expressions with a more generic property, e.g., NCnc subsuming LNCnc
Exception: Ratio in LOINC is a sibling to MRTO or SRTO and is interpreted as "other ratio." It is mapped to Ratio in SNOMED CT, left primitive and does not subsume expressions which include MRTO or SRTO.
There is no subsumption of expressions when the LOINC Terms contain Point in time or timed measurements.
3.5 Additional Information
Additional information is designated for each LOINC Part to SNOMED CT value/attribute map:
content origin (LOINC or SNOMED CT or both)
correlation (exact, narrower, broader)
attribute (where a Part is used in an expression)
refset identifier
module identifier
effective time of map
map status for active vs inactive
unique identifier of map row (GUID)
Additional information is designated for each LOINC Term to SNOMED CT post coordinated expression:
content origin (LOINC or SNOMED CT or both)
correlation (exact, narrower, broader)
definition status (primitive or fully defined)
refset identifier
module identifier
effective time of expression
expression status for active vs inactive
unique identifier of map row (GUID)
Some SNOMED CT Concepts included in the file do not specifically map to one LOINC Part, e.g. some specimen concepts and some technique concepts
Disjunctive substance LOINC Parts are not included in this Production file release because we do not have a way to model them in SNOMED CT at this time. Therefore they are on hold until a solution is provided. These LOINC Parts were included in previous Alpha releases but have been removed from the Production Release as a result of this recent decision.
Active, discouraged and trial LOINC Terms are included in the mapping as some implementers are using these LOINC Terms. Deprecated LOINC Terms and Parts are not included in the mapping.
This file contains some LOINC Parts mapped to the same SNOMED CT concept which are being reviewed by Regenstrief to see if they are in fact duplicates
Example: 11-Ketoandrosterone and 11-Oxoandrosterone are both mapped to 57244003 | 11-Ketoandrosterone (substance)
Example: Macroglobulin and IgM are both mapped to
This file contains some duplicate LOINC Parts which RII is planning on deprecating soon.
Example: E coli shiga-like and verotoxin are equivalent. RII indicated that they will deprecate the verotoxic term and map it to the shiga-like term.
Some naming conventions in SNOMED CT concepts are not consistent
Example: "Immunoglobulin G antibody to Yersinia" vs "Immunoglobulin A antibody to Yersinia species"
SNOMED CT utilizes multiple descriptions per concept, and some of the descriptions used may not be common parlance in laboratory medicine.
Example: The LOINC Part for “Bacteria identified” is mapped to Superkingdom Bacteria (organism) in SNOMED CT. We agree that the fully specified SNOMED CT name for this concept is not used in most lab reports; the concept it signifies (“Bacteria”) is used in lab reports. This concept has a description/synonym of “Bacteria” in SNOMED CT.
Example: The LOINC Part for “Fungus identified” Is mapped to Kingdom Fungi (organism) in SNOMED CT. This concept currently has a description of “Fungi.” Although another SNOMED CT - Fungus (organism) - would appear to be a more accurate match, there has been a recent recommendation by the Organism and Infectious Disease Model Project Group to retire 23496000 | Fungus (organism) | as a duplicate of 414561005 | Kingdom Fungi (organism) |. When this occurs we will add a description/synonym of “Fungus” to this concept.
The SNOMED International Microbiology Reporting Project Group discussed similar topics and posted guidelines in the SNOMED CT Editorial Guide in section 6.3.5. Observables and results for microbiology tests (https://confluence.ihtsdotools.org/x/fYu2AQ). These guidelines can be applied to Superkingdom Bacteria vs. Bacteria identified as well.
4 Quality Assurance
The files associated with this Production release were reviewed by a number of individuals including Terminologists at SNOMED International, Regenstrief Institute, and Linköping University. Quality assurance reviews were performed both manually and with some automated processes. Feedback received from reviewers (internal and external) on previous Alpha and Beta releases were considered and changes were implemented in this release where applicable. There are several known issues that are currently under review. Changes will be implemented when a resolution becomes available.
5 Next Steps
Next steps in the project include:
Finish the map of LOINC Parts to SNOMED CT concepts. Additional SNOMED CT concepts will need to be created to finish the map.
Use the above map to create more expressions for LOINC Terms.
Resolve and/or note content and modeling known and upcoming issues.
- Implement the SNOMED CT Observables model to define existing pre-coordinated SNOMED CT concepts.
6 Acknowledgements
We would like to acknowledge Mélissa Mary for her thorough review, analysis, and feedback on two previous versions of the file releases as well as for her presentation of her findings to the SNOMED International Content Committee in April, 2015.
We would like to acknowledge Bruce Goldberg and Michael Smith of Kaiser Permanente for their valuable assistance in early mapping in this project and for reviewing many versions of the expressions map before publication.
We would like to acknowledge Daniel Karlsson for his assistance with tooling, content decisions, and review during this release and previous releases.
We would like to thank all the reviewers who provided feedback on previous releases of this work.
7 References
Production release files of LOINC - SNOMED CT Cooperation Project are available at: https://mlds.ihtsdotools.org/#/ihtsdoReleases/ihtsdoRelease/50041
“Using LOINC with SNOMED CT:” http://snomed.org/snomedloinc
COOPERATION AGREEMENT dated July 2013 Between The International Health Terminology Standards Development Organisation (IHTSDO) and The Regenstrief Institute, Incorporated (RII): http://www.snomed.org/about/partnerships/loinc
Draft SNOMED CT Observables Model described in a PowerPoint presentation titled “Observables model in the context of the concept model:” https://confluence.ihtsdotools.org/x/spEeAQ
Note that viewing this presentation requires being a member of Observable and Investigation Model Project on Confluence. If you are not yet a user of this project on Confluence, please request addition through the site.
Observables and investigation procedures redesign Inception/Elaboration document: https://confluence.ihtsdotools.org/x/yY5UAQ
Note that viewing this presentation requires being a member of Observable and Investigation Model Project on Confluence. If you are not yet a user of this project on Confluence, please request addition through the site.
6.3.3 Attributes for observable entities and evaluation procedures in the SNOMED CT Editorial Guide: https://confluence.ihtsdotools.org/x/ZRn-AQ
Information about Expressions can be found in the “SNOMED CT Technical Implementation Guide:” https://confluence.ihtsdotools.org/x/4ICZAQ
"LOINC Users' Guide" available here: https://loinc.org/learn/
SNOMED International (formerly and also known as The International Health Terminology Standards Organization - IHTSDO): http://www.snomed.org/
SNOMED International Confluence: https://confluence.ihtsdotools.org/
Logical Identifiers Names and Codes (LOINC): http://loinc.org/
Approvals
Final Version | Date | Approver | Comments |
|---|---|---|---|
| 1.0 | Suzanne Santamaria | ||
| 1.0 | Lesley MacNeil |
Download .pdf here:
Draft Amendment History
Version | Date | Editor | Comments |
|---|---|---|---|
0.1 | 20170502 | Suzanne Santamaria | Initial draft |